Using protein dynamics to study role of water in biological reactions
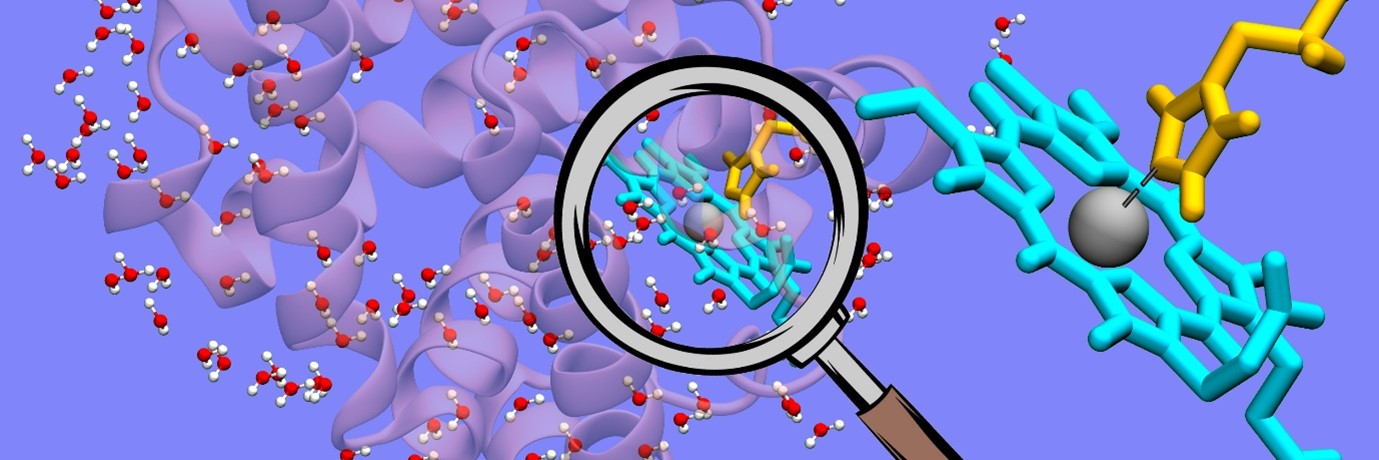
Water is regarded as the “matrix of life”. But how exactly does water help in biological reactions? This question has perplexed researchers for years. In recent work led by Prof Biman Bagchi at the Solid State and Structural Chemistry Unit (SSCU), researchers have attempted to understand this issue, by focusing on three different proteins: myoglobin, GB1 and dimeric insulin. In each of these systems, they considered a single reaction coordinate (hydrogen bond or coordination) and studied the effect of water on its dissociation. More specifically, they calculated the fiction components on the bond, separately from protein and water.
Their investigation reveals that the cross-correlation between the forces from these two domains results in a negative friction term at the reaction coordinate, which reduces the total friction and effectively increases the reaction rate. The force spectrum reveals a bimodal 1/f noise characteristic which suggests a synergistic relation between the fluctuations in protein and water, and reveals the non-Markovian nature of the problem. The action-at-a-distance of water on protein function and dynamics is shown to lead to a tug-of-war between internal (due to protein) and external (due to water) frictions, revealed clearly by considering the three different proteins. The researchers demonstrate that such out-of-phase behaviour in protein-water interactions, pointed out here for the first time, facilitates reactions through reduction of the friction on the reaction coordinate.
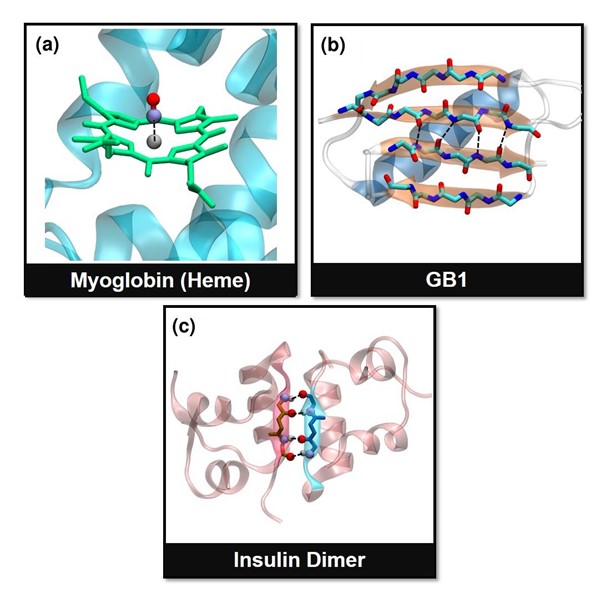
Figure: Pictorial description of the reaction coordinates in the three systems studied in this work. (a) Fe-CO bond in the heme unit of myoglobin, (b) hydrogen bonds (HBs) between parallel b-sheets in GB1, and (c) the junction HBs in insulin dimer. These bonds vectors act as the reaction coordinates (black dashed lines) in this study.
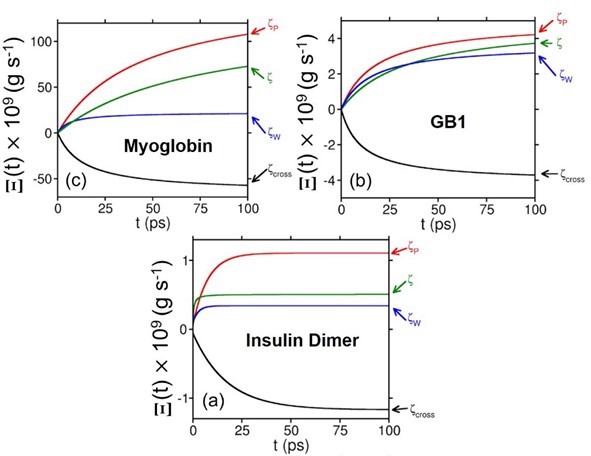
Figure: Time evolution of the auxiliary function (defined in the paper) for different contributions to the total friction experienced by (a) Fe-CO bond in the heme unit of myoglobin, (b) HBs between parallel b-sheets in GB1, and (c) the junction HBs in insulin dimer. Color code: Red, contributions from protein; blue, from water; and black, the cross term. The total response is represented by the green color. The cross term contributes a huge negative value, which brings down the value of total friction.
Reference:
S. Mukherjee, S. Mondal, S. Acharya, and B. Bagchi, Tug-of-War between Internal and External Frictions and Viscosity Dependence of Rate in Biological Reactions, Phys. Rev. Lett. 128, 108101 (2022).
https://doi.org/10.1103/PhysRevLett.128.108101
Group homepage:
https://sites.google.com/view/bimanbagchi/







