RNA granules: Tiny dots with big impact
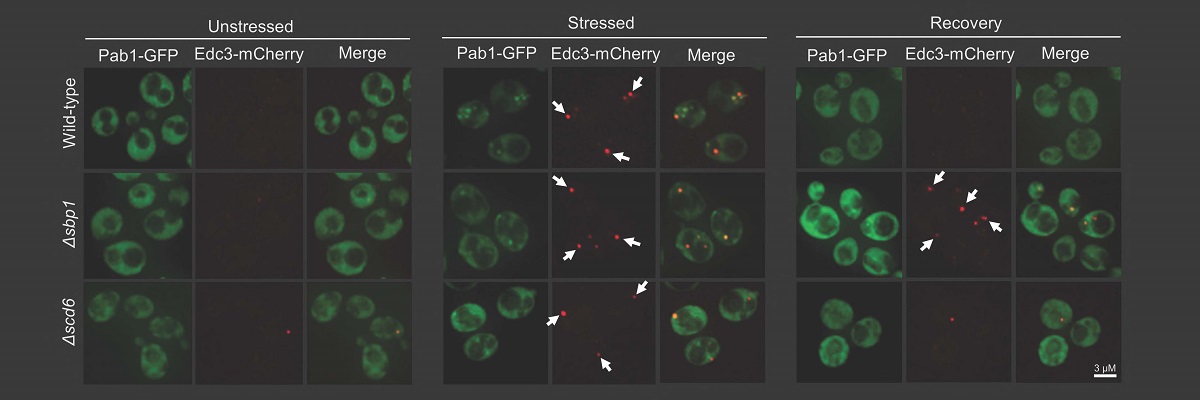
A recent study from the lab of Purusharth Rajyaguru, Associate Professor at the Department of Biochemistry, has identified a protein from the single-celled organism yeast with the ability to disassemble RNA granules such as Processing body (P-body). RNA granules are conserved membrane-less punctate structures that play an important role in determining the fate of messenger RNA (mRNA). Messenger RNAs, upon getting exported to the cytoplasm, can either be translated, remain stored in a silent state or get degraded. RNA granules play a key role in orchestrating these decisions which, in turn, is important in the context of health and diseases.
The study, published in Nature Communications, identifies the exciting role of the Sbp1 protein in disassembling RNA granules. Interestingly, this protein consists of repeats of arginine (R) and glycine (G) amino acids, known as the RGG-motif. Sequences with amino acid repeats are referred to as low complexity sequences as such sequences do not have representation of all amino acids. Such low complexity sequences have previously been implicated in the assembly of RNA granules, however, their ability to disassembly RNA granules was unknown. Sbp1 uses the low complexity RGG-motif sequence to disassemble RNA granules.
Although the study was performed using yeast, it also has implications for human disease biology. Protein aggregates in neurons are a hallmark in patients suffering from neurodegenerative disorders such as Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal dementia (FTD). These aggregates often contain proteins which are markers of RNA granules such as P-bodies and stress granules. Previous studies suggest that the inability of aberrant RNA granules to disassemble leads to accumulation of the aggregates which are a hallmark of neurodegenerative disorders. Therefore identifying and understanding mechanisms of granule disassembly will have therapeutic value.
Strikingly, these results indicate that Sbp1 (a yeast protein) manages to disassemble aggregates of EWSR1, a protein implicated in ALS. Therefore, Sbp1, through its RGG-motif, is not only important for disassembly of P-bodies in yeast but also aggregates of disease-relevant human proteins. The team is currently screening for more low complexity sequences with such activity. They will be testing the role of RGG-motif sequences in disassembling aggregates of disease-relevant proteins in neuronal cells and subsequently animal model systems.
This study, therefore, highlights the potential therapeutic role of the RGG-motif containing low complexity sequences in the context of neurodegenerative disorders.
REFERENCE:
Roy, R., Das, G., Kuttanda, I.A., Bhatter, N. and Rajyaguru, P.I., Low complexity RGG-motif sequence is required for Processing body (P-body) disassembly. Nat Commun 13, 2077 (2022).
https://www.nature.com/articles/s41467-022-29715-5
LAB WEBSITE:
https://rajgodhuli.wixsite.com/rajyaguru-lab






