Harnessing the power of AI and Raman spectroscopy to identify disease-causing bacteria
– Shrivallabh Deshpande
—————
Rapidly identifying the causative bacterium can help clinicians treat and manage the symptoms of infectious diseases more efficiently. Current gold-standard tests are either time-consuming – like culture tests – or work only for a specific type of sample – nucleic acid amplification-based methods are time-intensive and involve bacterial lysing.
For several years, researchers from IISc, led by Siva Umapathy, have been working on combining Raman spectroscopy with AI for applications in diagnostics and healthcare. They have previously shown how the combination can be used to detect COVID-19 biomarkers in blood samples – an accomplishment that won the Challenger award at the NASSCOM AI Game Changer Awards Programme last year.
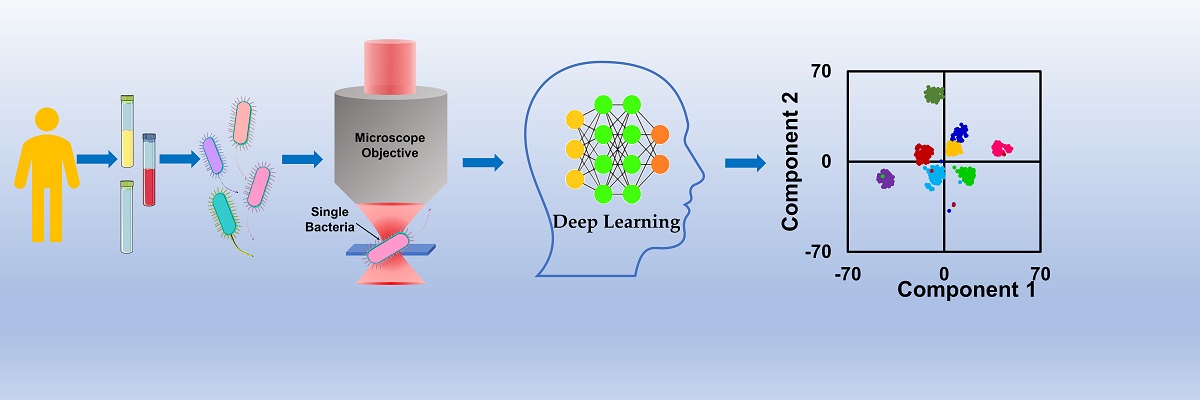
In a new study, Siva Umapathy’s and Deepak Kumar Saini’s teams demonstrate yet another application for this combination: quickly identifying bacterial pathogens in different types of clinical samples.
In Raman microspectroscopy, a laser beam is flashed on the sample using an optical microscope, and the light scattered by the bacterium gives a unique spectrum, which would allow the scientist or clinician to identify them individually. But making out subtle differences between different spectra can be challenging for the human eye. This is where AI can help.
The researchers employed a state-of-the-art Deep Learning model called Residual Network (ResNet) to identify and differentiate between the spectra of eight bacterial species widely implicated in hospital-acquired infections, famously called the ESKAPE pathogens. The samples were processed and procured by the team led by Dr Sneha Chunchanur at the Bangalore Medical College and Research Institute, and were subjected to Raman spectra collection without culturing. Using an approach called transfer learning, the researchers were able to train the model using very limited clinical data; the model was then able to identify the diverse bacterial species with an accuracy of 99.99%, according to the authors.
Such studies show how using AI can help cut down the time involved in diagnosis and speed up the treatment and management of bacterial infections.
REFERENCE:
Singh S, Kumbhar D, Reghu D, Venugopal SJ, Rekha PT, Mohandas S, Rao S, Rangaiah A, Chunchanur SK, Saini DK, Umapathy S, Culture-Independent Raman Spectroscopic Identification of Bacterial Pathogens from Clinical Samples Using Deep Transfer Learning, Analytical Chemistry (2022).
https://doi.org/10.1021/acs.analchem.2c03391
LAB WEBSITES:
Deepak Kumar Saini – https://sites.google.com/site/sainislab/
Siva Umapathy – https://ipc.iisc.ac.in/~su/




