Multiscale digital twins and battles of mutant programs to study cancer
Tissues and organs form when cells exchange molecular signals, interact with their surroundings, and move around to organise themselves based on principles of physics and chemistry. Such organisational principles vary along evolutionary scales to give rise to diverse organs and tissues seen within the animal kingdom. These principles are also relevant to understanding the changes that occur within organs in diseases such as cancer, where aberrant changes in genes alter behaviours of cells, leading to the disruption of tissue organisation. Can the subsequent changes that occur within a tumour, such as its spread, be understood through the lens of the same principles that gave rise to the organ?
In a paper published in the Journal of Theoretical Biology, Durjay Pramanik, Mohit Kumar Jolly and Ramray Bhat from the Centre for BioSystems Science and Engineering (BSSE) and the Department of Molecular Reproduction, Development and Genetics (MRDG), IISc, recreated the breast cancer microenvironment within a computational framework. By accounting for diverse interactions between cancer cells and their surrounding matrix and using clustering approaches, the team showed that the complexity of the matrix contributes crucially to determining whether cancer cells spread out of their original focus as single cells, as collectives, or through exotic combinations of the two. This choice has deep consequences for how fast cancer metastasizes and how efficient therapeutic outcomes are.
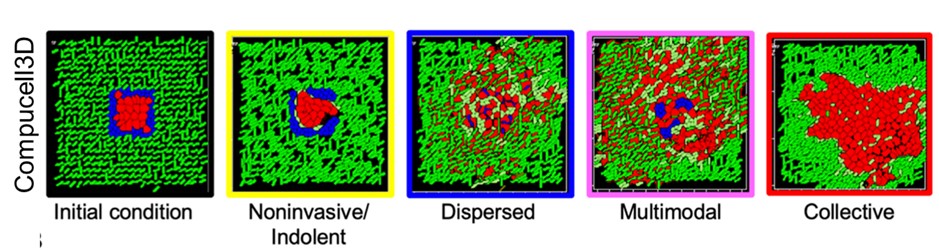
Given that tumours are populations of cells with varying phenotypes and hence fitnesses, competing for shared space and resources, they could also be studied on an evolutionary time scale. This led Bhaskar Kumawat and Ramray Bhat to set up evolutionary simulation experiments of digital organisms ‒ individual computer programs with a genome-like instruction template, which are capable of replication, metabolism and cooperative behaviour, but also susceptible to genomic alteration very much like cancer cells. The researchers found that, under high mutation conditions, discrete populations evolved to choose between existing either as a fast-dividing but genetically similar population of organisms with no collective behaviour, or as a relatively slower-dividing but genetically diverse population which could cooperate to share metabolic resources for survival. The choice between the two strategies depended on permitted population size and resources. This study was published in the BMC Ecology and Evolution.
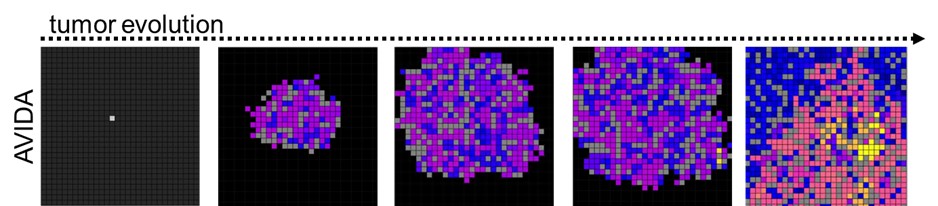
These studies suggest how generic interactional principles can on their own (and especially if given sufficient time and space) contribute to startlingly complex emergent behaviors seen in cancer such as heterogeneity and collective invasion. They represent fresh forays into a theoretical understanding of the disease. Of note, the first authors of both studies were undergraduate students of biology at IISc.
REFERENCES:
Pramanik D, Jolly MK, Bhat R. Matrix adhesion and remodeling diversifies modes of cancer invasion across spatial scales. J Theor Biol. 2021 Sep 7;524:110733. doi: 10.1016/j.jtbi.2021.110733. Epub 2021 Apr 30. PMID: 33933478.
Kumawat B, Bhat R. An interplay of resource availability, population size and mutation rate potentiates the evolution of metabolic signaling. BMC Ecol Evol. 2021 Apr 7;21(1):52. doi: 10.1186/s12862-021-01782-0. PMID: 33827412; PMCID: PMC8028831.
LAB WEBSITES:
https://morphogenesisiisc.wixsite.com/home
https://mkjolly15.wixsite.com/cancersystemsbiology





