Using nanosheets to identify bacterial strain and drug resistance
– Sindhu M
Microorganisms like bacteria cause many infectious diseases, which are generally treated with antibiotics. The causative microbe must first be identified to prescribe the appropriate antibiotic. Researchers from the Department of Organic Chemistry and Department of Molecular Reproduction, Development and Genetics in IISc have developed an optical nanosheet sensor to identify these disease-causing microbes from urine samples.
Optical sensors are faster and simpler than culture-based methods and PCR. Nanosheet sensors such as those made of Graphene Oxide (GO) have a high surface area to volume ratio, but have been largely unsuitable for biological applications so far because of their poor stability in aqueous media such as urine or blood.
To overcome these limitations, the researchers designed chemically modified Molybdenum Disulfide (MoS2) nanosheets, which are water soluble and positively charged, and bind to the negatively charged cell membrane of microbes. They coupled Green Fluorescent Protein (GFP) to the nanosheet to visualise the interaction between the microbe and the nanosheets. The difference in intensity of the GFP signal after adding the microbe sample was used to differentiate bacterial strains.
The sensor can also be used to identify whether the strain is resistant to antibiotics or not. Antibiotic-resistant bacteria have altered membrane properties to reduce the intake and increase the expulsion of the antibiotic. Thus, they interact differently with the nanosheet. The researchers say that the sensor can be also used to study the effect of drugs on diseased cells.
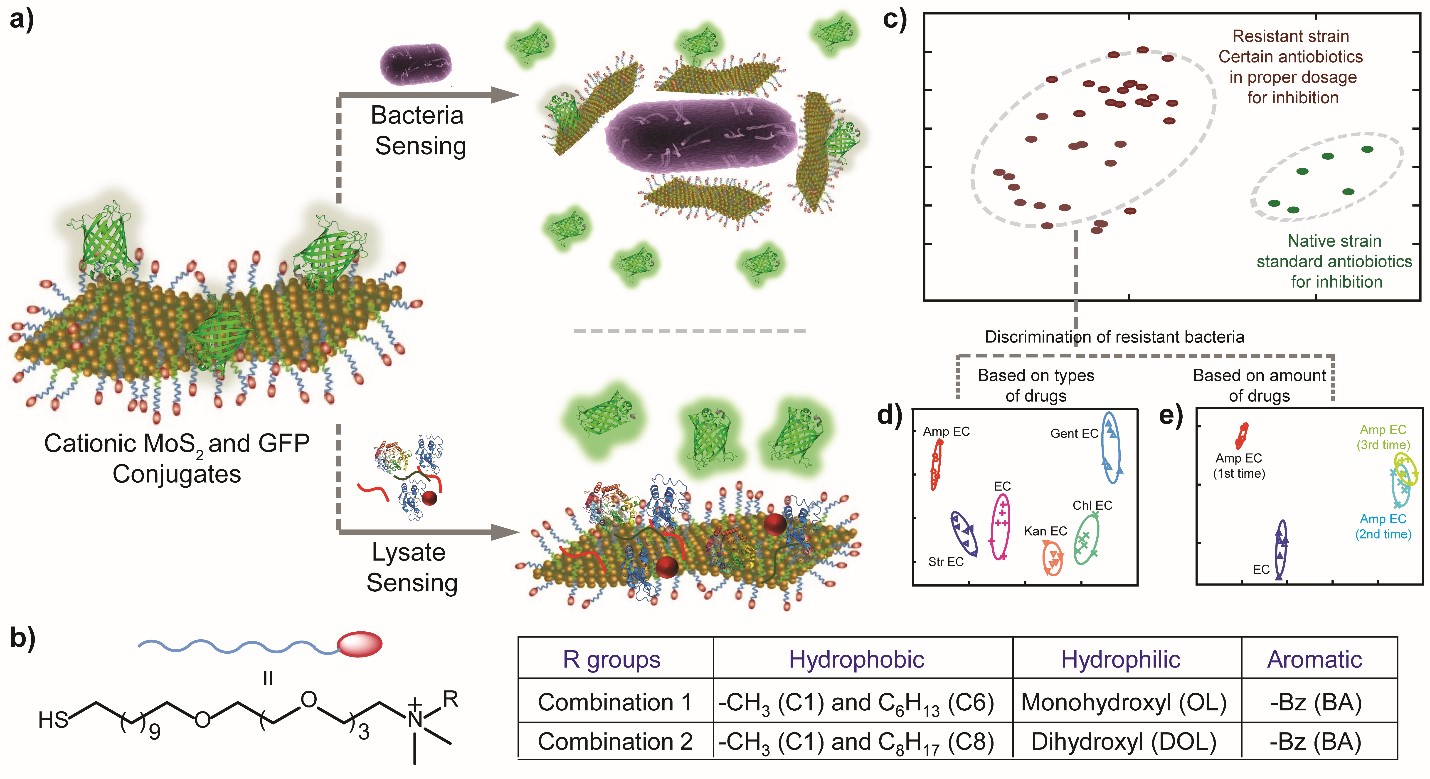
a) Schematic representation of the sensing mechanism for bacteria and their lysates. b) Structure of cationic thiol ligands with different head groups at the non-thiol end. c) Canonical plots showing differentiation of discrimination of the native strain from resistant ones through both lysate and whole-bacteria sensing. The resistant bacteria were classified based on d) types and e) amounts of drugs through statistical output.
Reference:
Pradipta Behera, Krishna Kumar Singh, Deepak Kumar Saini, and Mrinmoy De, Rapid discrimination of bacterial drug resistivity by array based cross-validation using 2D-MoS2. Chem. Eur. J. (2022).
Doi: https://doi.org/10.1002/chem.202201386
Lab websites:
Mrinmoy De: https://orgchem.iisc.ac.in/mrinmoy_de/
Deepak Kumar Saini: https://sites.google.com/site/sainislab/home







