To Be, or Knot To Be: RNA Pseudoknots Allow Toxin-Antitoxin Assembly
– Kredai Raaman
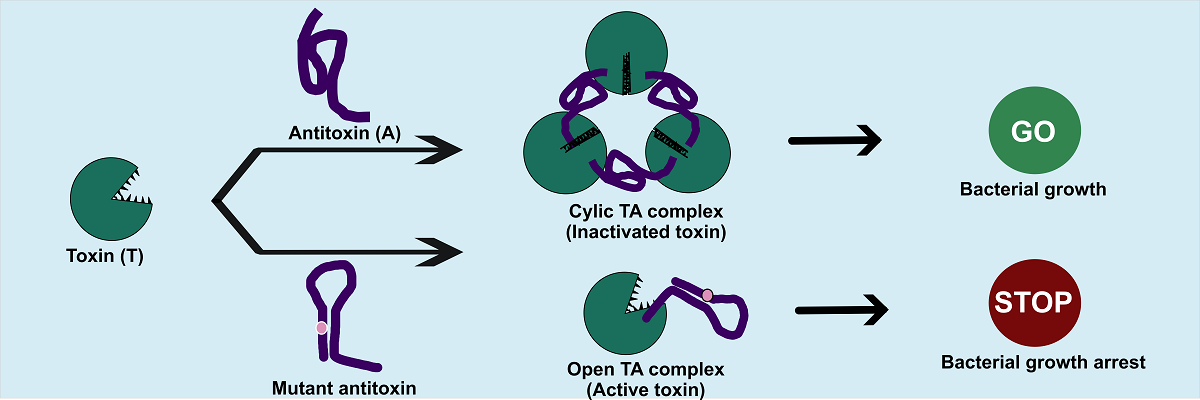
The antitoxin pseudoknot is crucial for the assembly of the toxin-antitoxin complex and for inhibiting the toxin (Image courtesy: Harshita Dutta and Mahavir Singh)
Found almost universally in bacteria, toxin-antitoxin systems are genetic modules that pair a destructive toxin with a neutralising antitoxin in a delicate balance of life and death. These toxins play an essential role in the bacterial defence against viruses, but in healthy bacteria, they must be bound and kept inactive within toxin-antitoxin complexes to prevent cell damage.
In a recent study, researchers from Mahavir Singh’s lab at the Molecular Biophysics Unit (MBU) focused on a class of systems where the antitoxin is a small RNA molecule with a pseudoknot (a feature formed by a pair of interwoven stem-and-loop structures).
The team examined the importance of these pseudoknots in the assembly of toxin-antitoxin complexes. They generated mutant antitoxins by altering single nucleotides (the building blocks of nucleic acids) at various positions in the RNA sequence and tested them with toxin-producing E. coli cells. They found that E. coli cells with some of the mutations in the antitoxin’s pseudoknot region did not survive. Thus, they concluded that the pseudoknot plays a critical role in toxin-antitoxin assembly and inhibition of the toxin.
Normally, three antitoxin units bind three toxin units, forming a ring-like complex that keeps the toxins locked up. Pseudoknot mutants, however, had significantly altered structures – though they could still bind toxins, they failed to form the strong, cyclic complexes required for effective neutralisation. Instead, they formed fragile, open complexes from which the toxins could easily escape. Further tests showed that these pseudoknot mutants were less stable, unfolded more readily, and bound the toxin with less than one-hundredth the strength of unmutated antitoxins.
These results highlight the delicate precision of RNA folding. For bacteria, even single-nucleotide errors in the pseudoknot can mean self-destruction, illustrating the importance of sequence to structure and structure to function. Further, these results suggest that molecules that can target and destabilise these pseudoknots would be good candidates for new antibacterial drugs and allow us to use the bacteria’s own defence mechanisms against them.
REFERENCE:
Dutta H, Manikandan P, Singh M, Role of Antitoxin RNA Pseudoknot in Regulating Toxin Activity and Toxin-antitoxin RNP Complex Assembly, Journal of Molecular Biology (2025).
https://www.sciencedirect.com/science/article/pii/S0022283625004760
LAB WEBSITE:
https://singhmlab.weebly.com/




