New insights into stem cell reprogramming
– Shloak Vatsal
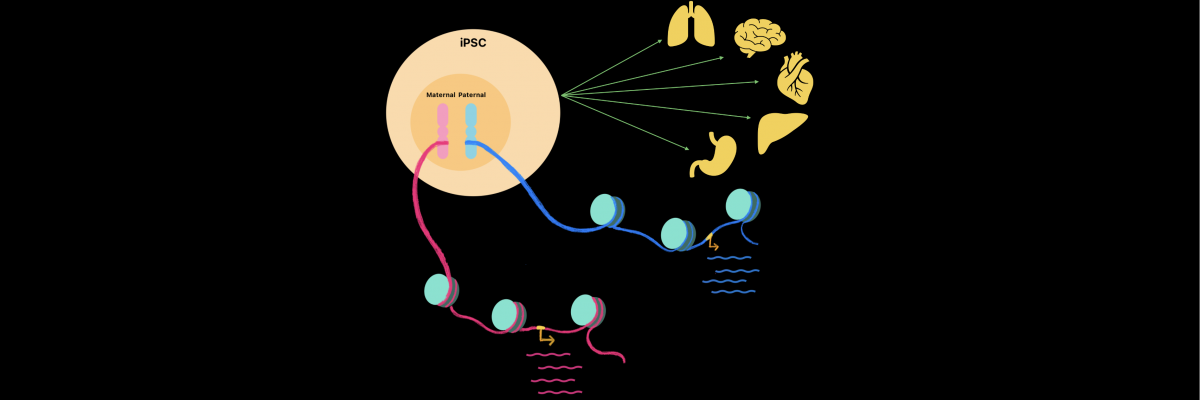 Image: Swati Negi and Akash Kalita
Image: Swati Negi and Akash Kalita
Different cell types from the adult body can be reprogrammed into induced pluripotent stem cells (iPSCs). Interestingly, iPSCs can give rise to different tissues and organs and serve as an excellent model to study early development and cell fate specification. Hence, in recent years, the molecular mechanisms of iPSC reprogramming have gained immense interest. It is known that the precise expression of genes contributes to iPSC reprogramming. However, how the individual allele (paternal and maternal copy) of a gene contributes to fine-tuning the expression of genes involved in iPSC reprogramming remains unknown. Interestingly, the two alleles of a gene can transcribe through sporadic bursts to produce mRNA either independently or co-ordinately, giving rise to allele-specific transcriptional bursting.
The IISc team, led by Srimonta Gayen, Assistant Professor at the Department of Developmental Biology and Genetics, profiled the transcriptional burst kinetics of individual alleles of genes. They used single-cell RNA-sequencing analysis of different stages of reprogramming of mouse embryonic fibroblasts (MEF) to iPSC.
The team found that many genes related to iPSC reprogramming pathways exhibit burst-like expression whose frequency and size are similar between the respective alleles. However, they found that while most of the genes exhibited semi-coordinated allelic bursting, only some showed highly co-ordinated allelic bursting (both of the alleles bursting at the same time), while others showed independent bursting. They then demonstrated that highly coordinated genes have higher expression levels compared to the semi-coordinated and independent genes. Also, allelic bursting in many genes crucial to iPSC reprogramming – such as those involved in translation, protein folding, nuclear pore formation, pluripotency maintenance and so on – occurs in a highly coordinated fashion. This suggests that coordination between the two alleles fine-tunes the amount of gene expression to drive precise reprogramming.
The team also showed that epigenomic states can drive this coordination by controlling the DNA transcribing machinery’s access to the chromatin, the structure within which DNA is tightly packed inside the nucleus. They found that genes undergoing highly coordinated bursting have similar access between alleles, whereas alleles of semi-coordinated or independent genes have differential levels of access. This controls how much of the transcribing machinery binds to the genes and to what levels the genes are expressed. The team also found that highly coordinated genes are enriched with important chromatin accessibility factors, such as H3.3, a protein whose role in bursting they have reported for the first time.
Altogether, the study provides insights into how paternal and maternal copies of individual genes join hands together to fine-tune gene expression during iPSC reprogramming.
REFERENCE:
Ayyamperumal P, Naik HC, Naskar AJ, Bammidi LS, Gayen S, Epigenomic states contribute to coordinated allelic transcriptional bursting in iPSC reprogramming, Life Science Alliance (2024)
http://doi.org/10.26508/lsa.202302337
LAB WEBSITE:
https://srimontasd.wixsite.com/gayen-lab



